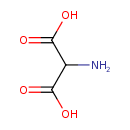
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-001i-2910000000-56ddccdbe51afd85dd6c | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-0gb9-1790000000-1a2cf2f982d08de4d2c9 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-001i-2910000000-56ddccdbe51afd85dd6c | Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0gb9-1790000000-1a2cf2f982d08de4d2c9 | Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0012-1920000000-706a1afd4a87657ef924 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00xu-9200000000-a92a1873b06d13a3cb1b | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-006t-7910000000-820eb39fc624884c77c9 | Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-05i0-9000000000-6da4c50b66acac061cd8 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-081c80e5b8aaa8f7ba34 | Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-00fr-9000000000-4dbbb4215703aa499e1b | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-8900000000-ce915814c1e82c9da3e2 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9200000000-c4dc8ccb28224391491f | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-9000000000-dda49c2194c794dfeebb | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-2900000000-b31a9403ea30555205e6 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01b9-5900000000-8f53ff3974857296fab8 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000000000-179c8f28fed467f08059 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-9200000000-2cbb45bb0d131a8a5a4c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-6bf2575f705bcef140f5 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-1f92eb692a3016f51886 | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-3900000000-2c6e9d09fa238ac1618c | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9400000000-ce7be9ee668d87a767bd | Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dl-9000000000-a28af5b62f46a4285ac2 | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 1H NMR Spectrum | Not Available | Spectrum |
| 1D NMR | 13C NMR Spectrum | Not Available | Spectrum |