| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-07-13 19:52:36 UTC |
|---|
| Update Date | 2016-07-20 21:04:15 UTC |
|---|
| Lmdb | LMDB00562 |
|---|
| Secondary Accession Numbers | None |
|---|
| Metabolite Identification |
|---|
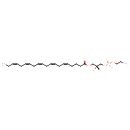
| Common Name | LysoPE(20:5(5Z,8Z,11Z,14Z,17Z)/0:0) |
|---|
| Description | LysoPE(20:5(5Z,8Z,11Z,14Z,17Z)/0:0) is a lysophosphatidylethanolamine or a lysophospholipid. The term 'lysophospholipid' (LPL) refers to any phospholipid that is missing one of its two O-acyl chains. Thus, LPLs have a free alcohol in either the sn-1 or sn-2 position. The prefix 'lyso-' comes from the fact that lysophospholipids were originally found to be hemolytic however it is now used to refer generally to phospholipids missing an acyl chain. LPLs are usually the result of phospholipase A-type enzymatic activity on regular phospholipids such as phosphatidylcholine or phosphatidic acid, although they can also be generated by the acylation of glycerophospholipids or the phosphorylation of monoacylglycerols. Some LPLs serve important signaling functions such as lysophosphatidic acid. Lysophosphatidylethanolamines (LPEs) can function as plant growth regulators with several diverse uses. (LPEs) are approved for outdoor agricultural use to accelerate ripening and improve the quality of fresh produce. They are also approved for indoor use to preserve stored crops and commercial cut flowers. As a breakdown product of phosphatidylethanolamine (PE), LPE is present in cells of all organisms. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (5Z,8Z,11Z,14Z,17Z-Eicosapentaenoyl)-lysophosphatidylethanolamine | HMDB | | 1-Eicosapentaenoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | HMDB | | LPE(20:5) | HMDB | | LPE(20:5/0:0) | HMDB | | LPE(20:5n3/0:0) | HMDB | | LPE(20:5W3/0:0) | HMDB | | Lyso-pe(20:5) | HMDB | | Lyso-pe(20:5/0:0) | HMDB | | Lyso-pe(20:5n3/0:0) | HMDB | | Lyso-pe(20:5W3/0:0) | HMDB | | LysoPE(20:5) | HMDB | | LysoPE(20:5/0:0) | HMDB | | LysoPE(20:5n3/0:0) | HMDB | | LysoPE(20:5W3/0:0) | HMDB | | Lysophosphatidylethanolamine(20:5) | HMDB | | Lysophosphatidylethanolamine(20:5/0:0) | HMDB | | Lysophosphatidylethanolamine(20:5n3/0:0) | HMDB | | Lysophosphatidylethanolamine(20:5W3/0:0) | HMDB | | 1-(5Z,8Z,11Z,14Z,17Z-Eicosapentaenoyl)-2-hydroxy-sn-glycero-3-phosphoethanolamine | HMDB | | LysoPE(20:5(5Z,8Z,11Z,14Z,17Z)/0:0) | Lipid Annotator |
|
|---|
| Chemical Formula | C25H42NO7P |
|---|
| Average Molecular Weight | 499.5772 |
|---|
| Monoisotopic Molecular Weight | 499.269889215 |
|---|
| IUPAC Name | (2-aminoethoxy)[(2R)-2-hydroxy-3-[(5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoyloxy]propoxy]phosphinic acid |
|---|
| Traditional Name | 2-aminoethoxy(2R)-2-hydroxy-3-[(5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoyloxy]propoxyphosphinic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@@](O)(COC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CC)COP(O)(=O)OCCN |
|---|
| InChI Identifier | InChI=1S/C25H42NO7P/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-25(28)31-22-24(27)23-33-34(29,30)32-21-20-26/h3-4,6-7,9-10,12-13,15-16,24,27H,2,5,8,11,14,17-23,26H2,1H3,(H,29,30)/b4-3-,7-6-,10-9-,13-12-,16-15-/t24-/m1/s1 |
|---|
| InChI Key | KCWGBXXAJHVZNN-KOYQJJOGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-acyl-sn-glycero-3-phosphoethanolamines. These are glycerophoethanolamines in which the glycerol is esterified with a fatty acid at O-1 position, and linked at position 3 to a phosphoethanolamine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoethanolamines |
|---|
| Direct Parent | 1-acyl-sn-glycero-3-phosphoethanolamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-monoacyl-sn-glycero-3-phosphoethanolamine
- Phosphoethanolamine
- Fatty acid ester
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Amino acid or derivatives
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organonitrogen compound
- Hydrocarbon derivative
- Alcohol
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organooxygen compound
- Primary amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zfu-1930200000-60da6e8db97ffe90bc7c | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0200-4190030000-842d23f1219e1cbf7cc3 | Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9111110000-c677dd8a063d60341732 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9121100000-1017d05e20aaf7e01d2e | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9031000000-6ab784485e80d3e0ce99 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0uea-2367900000-f5d5530e332228c4268c | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ugl-9667100000-c16dfd1fb37f4a553ad8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-9100000000-0631e5e0ee8423df50ed | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-6009760000-3da1281c8765436b26c7 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9104100000-d726384503aa2b55adc6 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9741000000-595617d9ed40e3ec17a8 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-430b8d570b93b9965636 | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-5634900000-a52b5cde3af4d3eb83bf | Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-3926100000-fc43aefec6cceb097737 | Spectrum |
|
|---|